Reports of research work funded by grants prior to 2015
Victoria University of Wellington
Development of enzymatic tools to improve zebrafish models for metabolic disease and stem cell based regenerative medicine
EM Williams, DF Ackerley
School of Biological Sciences
This research aims to engineer pairs of nitroreductases such that each enzyme in the pair is specific for a particular prodrug substrate, ie unable to activate the prodrug recognised by the other enzyme in the pair. These nitroreductases will have utility in small animal cellular ablation studies (in particular zebrafish), where the animals have been engineered to express each nitroreductase in a different tissue type. Upon administration of one (or both) of the prodrugs, the target tissue(s) will be ablated without harm to surrounding cells. In a hypothetical example, one nitroreductase could be placed under genetic control of an insulin promoter and one under control of a glucagon promoter, enabling study of the effects of specifically ablating the α- or β-cells of the pancreas at a precise stage of development. Each engineered enzyme will also be partnered with a specific masked fluorophore to enable imaging of the nitroreductase-expressing cells prior to ablation.
This enzyme engineering study will have the following Specific Objectives:
- To use directed evolution to develop pairs of enzymes with non-overlapping specificities for different nitro-prodrug antibiotic substrates.
- To partner the evolved enzymes with fluorogenic molecules that can report specifically and effectively on their expression levels in transfected cell lines.
- To demonstrate the potential for multiplexed cellular ablation in mixed cell culture assays.
Genes encoding the evolved enzymes will then be sent to our collaborator, Dr Jeff Mumm at Johns Hopkins University, for proof-of-principle testing and application in zebrafish models of stem cell based regenerative medicine
Review of Specific Objectives and Outcomes
Specific Objective 1: To use directed evolution to develop pairs of enzymes with non-overlapping specificities for different nitro-prodrug antibiotic substrates
This was the major objective of the project. In brief, engineering of pairs of enzymes to induce them to each have strong preferences for different prodrug substrates was successfully achieved, thereby validating the experimental design and underlying concept of the project. However, it is likely that further engineering will be required to achieve a sufficient “therapeutic index” (ie enzymes that act upon an appropriate range of prodrug concentrations, such that each prodrug is only activated efficiently by one of the two engineered enzymes) to enable precise ablation of each of two different target tissues within a single model organism.
Enzymes were engineered as proposed in our original application; that is, by using a random mutagenesis strategy to mutate a gene that encoded a target nitroreductase enzyme, then cloning the resulting gene library into a plasmid and transforming E. coli cells with that library. To select mutated genes encoding an enzyme variant that had lost activity with the prodrug metronidazole the bacteria were plated on growth medium containing a high concentration of metronidazole (ensuring that the host cells could only grow into colonies if the nitroreductase variant they contained had lost the ability to activate metronidazole) as well as niclosamide (a nitro-compound that is toxic to bacteria unless nitro-reduced; ensuring that all colonies must still contain a generically active nitroreductase). All colonies were then counter-screened for activity with a second prodrug, eg tinidazole, to identify the variants that were still active with that second prodrug but had lost activity with metronidazole. Analysis of the top variant generated within the timeframe of this study is illustrated in Fig. 1. The process describe above was repeated in reverse (ie tinidazole included in the growth medium, followed by counter-screening for retention of activity with metronidazole) to generate nitroreductase variants that had an opposing specificity. This was achieved more easily, using as a start point a nitroreductase that already exhibited a discernible preference for metronidazole as a substrate. The activity profile of the top variant identified to date is illustrated in Fig. 2.
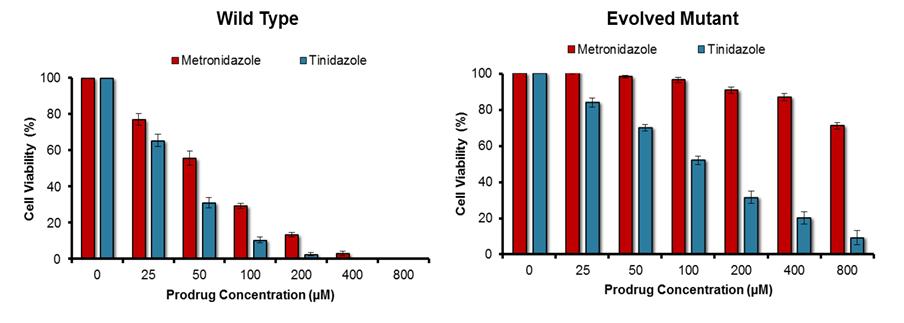
Fig. 1. Activity profile of non-engineered (“Wild Type”) and top engineered (“Evolved Mutant”) nitroreductase variants with metronidazole and tinidazole.
Each nitroreductase variant was expressed in Escherichia coli host cells, and the degree of sensitivity conferred to each prodrug was determined by replica culturing each strain in the presence of a dilution series of that prodrug. Decreased growth of the host strain is indicative of heightened activity with the prodrug substrate tested. Although the top evolved mutant had lost a degree of activity with tinidazole, it had lost far more activity with metronidazole, and was therefore strongly selective for tinidazole over metronidazole. N = 5 independent repeats, error bars indicate ± SEM.
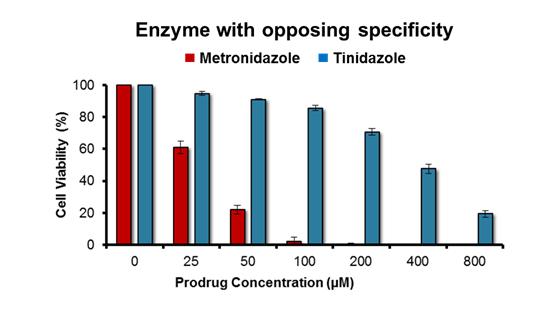
Fig. 2: Activity profile of top engineered nitroreductase variant with tinidazole and metronidazole.
The lead “metronidazole-favouring” variant identified to date exhibits a strong preference for metronidazole over tinidazole. N = 5 independent repeats, error bars indicate ± SEM.
Both metronidazole and tinidazole are 5-nitroimidazole prodrugs; that is, they share a similar core structure, making evolution of an enzyme that exhibits a strong preference for one substrate over the other a challenging task. Our results to date indicate that this is nevertheless a realistic goal, and the affordability and availability of these two prodrugs makes them attractive compounds to target. In ongoing work however, we have shown that more dramatic alterations in substrate specificity can likely be achieved when targeting a 5-nitroimidazole prodrug versus a 2-nitroimidazole prodrug, such as RSU-1069 (originally developed as a radiosensitizer).
Specific Objective 2: To partner the lead evolved enzymes with fluorogenic molecules that can report specifically and effectively on their expression levels in transfected cell lines.
We found that the lead “metronidazole favouring” nitroreductase variant (Fig. 2) exhibits strong activity with a red fluorogenic substrate named FSL178, which the wild type nitroreductase we selected to engineer is not able to activate. Disappointingly however, our lead “tinidazole-favouring” variant (Fig. 1, right panel) had lost all activity with each of the fluorogenic probes we tested. Thus, we will be unable to use these molecules to report on nitroreductase activity in transfected cell lines. This will not however restrict their ability to be used as ablation tools in transgenic animal models via the established method of co-expressing a prodrug-sensitising nitroreductase gene with a transcriptionally fused fluorescent reporter gene.
Specific Objective 3: To demonstrate the potential for multiplexed cellular ablation in mixed cell culture assays
This work was delayed by cloning difficulties, however these have now been resolved and human (HCT-116 colon carcinoma) cells are currently being transfected with our lead nitroreductase genes.
Genes encoding the top evolved enzymes have also been sent to our collaborator, Dr Jeff Mumm at Johns Hopkins University, for proof-of-principle testing in zebrafish models.



