Reports of research work funded by grants prior to 2013
Malaghan Institute of Medical Research
Using recombinant TFAM to transfect the mitochondria of rho zero cells
J W Baty
Background and objectives
Despite nuclear DNA transfection methods existing for many years, there are still very limited options for manipulating the genome of mitochondria. This is partly due to the difficulty of targeting exogenous DNA to the mitochondria. The inability to alter mitochondrial genomes in cells has hindered research into diseases caused by mitochondrial DNA mutations. However, it has been reported that recombinant Mitochondrial Transcription Factor A (TFAM), when constructed with a mitochondrial targeting sequence, is capable of binding DNA molecules and transporting them into the mitochondrial matrix of intact cells. We are testing this technology by using recombinant mouse TFAM (rmTFAM) to transfect entire mitochondrial genomes into mouse cells that have been depleted of their mitochondrial DNA (rho zero cells, ρ0). Specifically, our objectives are to:
- Construct a functional rmTFAM with a mitochondrial targeting sequence.
- Use the rmTFAM to transport fluorescently-labelled mitochondrial genomes into the mitochondrial matrix of ρ0 mouse cells.
- Measure the success of mitochondrial transfection by detecting restoration of normal mitochondrial function in transfected ρ0 mouse cells.
Objective 1: Construct a functional rmTFAM with a mitochondrial targeting sequence
We designed an rmTFAM with a protein transduction domain and hemagglutinin epitope to traffic the protein across the cell membrane into the cytoplasm, and with the native TFAM mitochondrial localization signal to target the protein to the mitochondrial matrix. GenScript (NJ, USA) was commissioned to express and purify the recombinant protein, which was shipped in December 2012. Aliquots of the recombinant protein are stored at -80oC.
Objective 2: Use the recombinant mouse TFAM to transport fluorescently-labelled mitochondrial genomes into the mitochondrial matrix of rho zero mouse cells
To test the ability of the rmTFAM to transport mitochondrial genomes into cells, mitochondrial DNA (mtDNA) was extracted from B16 melanoma cells and labelled with a fluorescent tag (Cy3) and then incubated with rmTFAM for 30 min at 37oC to allow the protein to bind to the mtDNA-Cy3 cargo. The rmTFAM-mtDNA-Cy3 complex was then incubated with 4T1 mammary fat pad carcinoma ρ0 mouse cells for 5–24 h and the cells analysed by fluorescence microscopy. Initial experiments showed that the ratio of rmTFAM to mtDNA was too high resulting in precipitation of the complex so that it was not taken up the cells. However, when less rmTFAM was used precipitation did not occur and uptake of the complex was observed within 6 hours, with maximum levels observed at 24 hours and persisting out to 48 hours (Figure 1a). Cells were also stained with MitoTracker Green to determine if the rmTFAM-mtDNA-Cy3 complex colocalised with MitoTracker green-stained mitochondria (colocalisation was seen as yellow-orange specks in Figure 1b).
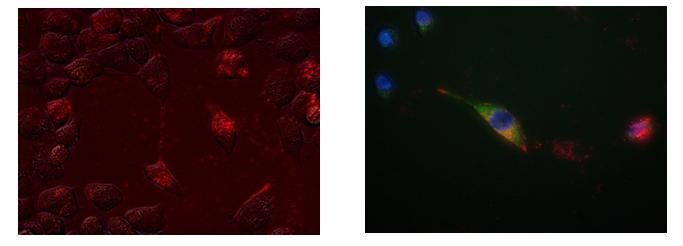
Figure 1: 4T1ρ0 (a) and 4T1 cells (b) treated with rmTFAM-mtDNA-Cy3. Cells were treated with rmTFAM- Cy3 complex for 24 h and then analysed by fluorescence microscopy to determine if the Cy3-labeled mtDNA entered the cells (Cy3 is observed as red/pink). (b) 4T1 cells were also stained with MitoTracker Green to determine if the Cy3-labelled mtDNA colocalised with the green-stained mitochondria. Blue indicates Hoesht 33342-counter stained nuclei of the 4T1 cells. (Both images are under 400x magnification).
Objective 3: Measure the success of mitochondrial transfection by detecting restoration of normal mitochondrial function in transfected ρ0 cells
To test the ability of rmTFAM-mtDNA to restore normal mitochondrial function to ρ0 cells, 4T1ρ0 cells were incubated with the complex for up to 24 hours, washed at least twice, and then incubated in medium that lacked pyruvate and uridine (supplements required by ρ0 cells for survival). Under the conditions tested so far, rmTFAM-mtDNA was unable to confer on 4T1ρ0 the ability to survive without pyruvate and uridine supplementation. One possibility is that the mitochondria of 4T1ρ0 are incapable of replicating the donor mtDNA therefore; we are also testing 4T1 cells that have had their endogenous mtDNA knocked-down by two-week treatment with dideoxycytidine. We hypothesize that the dideoxycitidine-treated 4T1 cells will still have functional mtDNA replicating machinery and more likely to be capable of using the exogenous mtDNA. To track uptake of B16 mtDNA in 4T1 cells we are using a signature sequence in the mitochondrial tRNA arginine (R) gene of B16 cells. We have shown that B16 cells are heteroplasmic for a sequence of 9/10 As in tRNA R, whereas 4T1 cells have only 8 As at the same position. Thus, successful uptake of B16 mtDNA by 4T1 cells will be indicated by the presence of 9/10 As in tRNA R. However, in experiments to date none of the rmTFAM-mtDNA-treated 4T1 cells have acquired the B16 tRNA R signature sequence.



