Reports of research work funded by grants prior to 2015
Victoria University of Wellington
Targeting Antibiotic Tolerance in Pseudomonas aeruginosa with DNA Aptamers
DJ Day and JP Soundy
School of Biological Sciences
The emergence of multi-drug resistant Gram-negative bacteria poses a significant threat to antibiotic therapy. The ongoing explosion of antibiotic resistant infections has been called a “ticking time bomb” with the “golden age of the antibiotic set to end unless action is taken”. This statement of doom and gloom by the United Kingdom’s Chief Medical Officer is a response to the global increase in antibiotic resistance, as the acquisition of resistance outpaces the discovery and development of new antibiotics.
Recent studies have shown that short DNA or RNA molecules termed aptamers can be selected, using a process called SELEX, from a large random library of sequences for their ability to bind a specific target. In some instances binding to the target can neutralise the biological function. In this project we have used the SELEX process to screen for aptamers able to bind the opportunistic pathogen Pseudomonas aeruginosa and are investigating whether aptamers can be used as a new type of antibiotic to inhibit bacterial growth. P. aeruginosa causes chronic infections in the lungs of cystic fibrosis that are highly resistant to antibiotic therapy, ultimately leading to premature death, as well as commonly infecting diabetic ulcers, where the bacteria form biofilms that impair healing causing significant morbidity.
Fig. 1 shows that successive rounds of SELEX selection results in enrichment of DNA libraries for sequences that are able to bind live P. aeruginosa, and that sequence in the seventh iteration are able to inhibit bacterial growth.
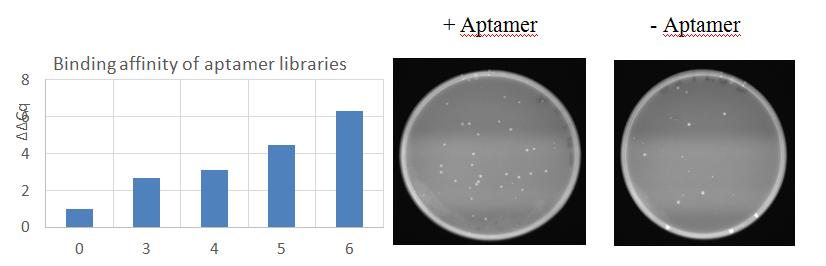
Fig. 1. SELEX selection increases the affinity of library pools for living PA isolated from biofilms. Incubation of biofilms cells with 500 nM of library L7 for 30 mins prior to plating results in decrease growth and fewer (50% reduction) in colony forming units.
Current experiments are using next generation sequencing of the libraries and bioinformatic analysis of the sequence data to identify enriched sequences (aptamers) that will be further characterised for putative antimicrobial activity.



