Reports of research work funded by grants prior to 2014
Institute of Environmental Science & Research Ltd
Validation of differential methylation at specific loci in adipose tissue before and after gastric bypass and the major weight-loss associated with it
D Macartney-Coxson*, M Benton, A Johnstone and R S Stubbs
Objective: Type-2 diabetes and obesity are diseases of epidemic proportions. Gastric bypass surgery results in major weight-loss and in many cases substantial amelioration of insulin resistance and type-2 diabetes. Using gastric bypass as a model system we have identified differentially methylated loci in adipose tissue before and after surgery. We wish to validate a number of these loci as a first step towards understanding their significance in disease.
Background: We previously investigated DNA methylation, in two different adipose tissues subcutaneous adipose and omentum before and after gastric bypass and associated weight-loss using the high-density Illumina 450K platform. A paired analyses before surgery and after subsequent weight-loss revealed significant differential methylation (Bonferroni p<1x10-7) in subcutaneous adipose and omentum at 3601 and 15 cytosine guanine dinucleotides (CpGs) respectively. This differential methylation was observed within obesity candidate genes, genomic regions associated with obesity, non-coding RNA loci and genes involved in epigenetic regulation and development. We sought WMRF funding to validate a number of these differentially methylated loci using an independent technique, pyrosequencing.
Method: Genomic DNA samples were sent to a commercial provider, Exiqon (USA). All CpG probe sites passing Bonferroni correction within a gene of interest were interrogated. For subcutaneous adipose these were: CETP (cg03232842), IFFO1 (cg10838410); CTGF (cg00516030, cg17359975); CMIP (cg02573873, cg05438708, cg05705335, cg08344351), and KCNQ1 (cg03371125, cg04902871, cg10678459, cg14637411, cg19698309, cg19923326). For omentum: PARD3B (cg12157387); PLIN4 (cg01907005); MYO1C (cg06579248), and PDE7B (cg04627183). Depending on the assay, amplicon and sequence content additional CpG sites surrounding the loci of interest were also analysed. Pyrosequence assays were designed, optimised, performed and analysed by Exiqon; both pyrograms and assay result data were supplied. Data analyses were performed using R version 2.15.2 [114], Bioconductor [115] packages
Results: Pyrosequence analyses of adipose DNA samples from the 15 individuals included in the original 450K array experiment were conducted, along with additional subcutaneous adipose (n=9 male, n=3 female) and omentum (n=5 males, n=2 females) samples.
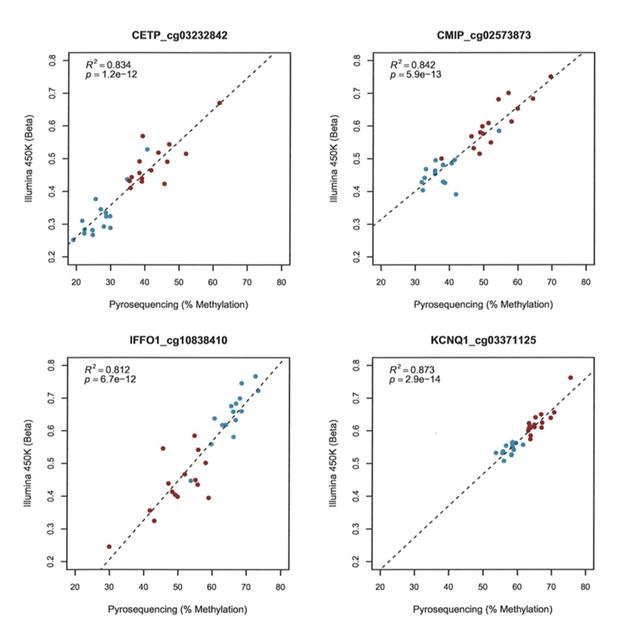
For the subcutaneous adipose analysis, loci were selected based on a number of criteria, a) most hyper- and hypo-methylated before and after weight-loss (CMIP, and IFFO1) b) genes implicated in obesity and/or type-2 diabetes (KCNQ1, CETP, CMIP, ADRAB2) and/or c) reported differential adipose mRNA expression in response to weight-loss/dietary intervention (CETP, CTGF). All CpG probe sites passing Bonferroni correction in a given gene were analysed (total 15 independent assays, see methods). First correlation analyses were carried out to compare the data from the two different assays (Illumina 450K and pyrosequencing). This revealed strong correlations for all sites (R2 =0.656-0.939, p=3.6x10-8 – 9.8x10-19). Figure 1 presents data for four representative CpG loci.
Analysis of the pyrosequence data for all samples from subcutaneous adipose revealed significant differential methylation of all 15 CpG sites interrogated in both females and males except 2 CpGs in KCNQ1 which were not differentially methylated in males. A comparison of methylation at each CpG between males and females revealed small but statistically significant differences (P<0.05) at a number of loci. Notably, six CpGs within an island in the CMIP loci all showed statistically more methylation in females than males after weight-loss as did three adjacent CpG sites in one the CETP gene.
For omentum we selected sites in the four genes with known or potential roles in obesity MYO1C, PLIN4, PARD3 and PDE7B. Significant correlations between the methylation assays was seen for PDE7B (R2 =0.671, p=1.9x10-8), PLIN4 (R2 =0.399, p=0.0001), and PARD3B (R2 =0.528 p=2.4x10-6). Differential methylation of MYO1C was not validated (R2 = -0.02 p=0.49).
Analysis of the data for all omentum samples showed differential methylation within PARD3B, PDE7B and PLIN4 before and after weight-lost in both males and females except 1 CpG site located 7 bases upstream of the transcription start site of PDE7B. None of the CpG loci within PARD3B, PDE7B and PLIN4 showed a significant difference in methylation level between males and females.
Increased DNA methylation within promoter regions is associated with transcriptional repression. The pyrosequence data for PLIN4 included six CpG sites immediately upstream of the transcription start site, therefore we averaged the percent methylation across this region and tested for a correlation (linear regression) with mRNA expression both before and after weight-loss. A strong correlation was observed after weight-loss (R2=0.55, p=0.0009) with less methylation corresponding to more mRNA expression. No correlation was observed before weight-loss (R2=-0.03, p=0.5). This contrasting observation may reflect a dysregulation of “normal” PLIN4 control in the obese omentum and is, to our knowledge, the first report for a potential role of DNA methylation in the regulation of obesity associated gene PLIN4.
In conclusion: We are very grateful to the WMRF for this funding because it has allowed us to robustly validate our initial observations of differential DNA methylation (with the Illumina 450K platform). Our adipose DNA methylation analyses, including the pyrosequence validation (funded by WMRF), has been presented in poster form at three conferences (copy emailed to WMRF 22/10/13) including one by Miles Benton (PhD student) at GeneMappers 2014 in Australia were he won the best poster prize. In addition, a manuscript has been submitted to Genome Biology. When a manuscript is accepted for publication a copy will be sent to WMRF.
Note: In addition, pyrosequencing analysis of two more genes involved in inflammation and obesity was performed on samples from subcutaneous adipose. These also showed an excellent correlation between the two different assays. These genes are the subject of on-going work with plans to draft a manuscript later this year.
Figure 1: Correlations between Illumina 450K array data and pyrosequence analysis. Representative data for a single CpG site in four genes is shown. Red indicates samples taken before and blue after gastric bypass. Illumina probe IDs are indicated after gene names.