Reports of research work funded by grants prior to 2012
Victoria University of Wellington
Environmental Survival of Neisseria meningitidis
C Swain and TW Jordan
School of Biological Sciences
The goal of this research is to identify proteins that vary among strains of meningococcal bacteria that have differing environmental survival. N. meningitidis (meningococcus), the causative agent of meningococcal disease, is a common commensal of the human nasopharynx. N. meningitidis is usually non-invasive and co-exists harmlessly with its human host, but in a small percentage of cases meningococci invade the lining of the throat and enter the bloodstream to cause meningococcal disease. It is generally considered that meningococci do not survive outside of the host, thus the most accepted route of transmission is via upper respiratory tract secretions and coughing and sneezing. Environmental transmission has been largely disregarded as a route for meningococcal disease due to the widely held belief that N. meningitidis is a temperamental organism. PhD student Claire Swain has shown, however, that some commonly carried strains of meningococcal bacteria can survive up to approximately 200 hours in the environment. Our current pilot study uses proteomic techniques to detect proteins differentially expressed between long-surviving and poor-surviving strains under different environmental conditions
Initially, the survival of 12 strains was examined under ambient conditions (16-28 oC and 28-64% relative humidity) during the course of a year. All isolates tested survived the process of desiccation. Initial percentages of survival depended on the isolate and ranged from 1 to 40%. All isolates tested could also survive the first four hours, but from that point onwards, survival of the different isolates varied substantially. Strains of the W:nt:P1.18-1,3 type had maximum survival times of around 24 hours, whereas strains of the B:4:P1.7-2,4 (the New Zealand epidemic strain type) and B:nt:P1.19-1,26 strain type (a common carriage strain in New Zealand) typically survived five days or more depending on environmental conditions. Meningococci strains were unable to survive more than 30 min at 36 oC/95% relative humidity. Subsequent testing was carried out at 30 oC and 22% or 30% relative humidity. Most strains survived longer at 22% relative humidity with the exception of two strains including H34.
N. meningitidis strains are characterised on the basis of their polysaccharide capsule and two outer membrane proteins called PorA and PorB. There are 13 different capsule types in meningococci and they are known as serogroups. Only 5 serogroups – A, B, C, W, and Y commonly cause disease. Survival ability is strongly linked to strain type. Poor survivors tend to be only of the type W:nt:P1.18-1,3, and strong-survivors tend to be of the type B:nt:P1.19-1,26. No W capsulated strain was able to survive well, and no B:nt:P1.19-1,26 strain survived poorly. Figure 1 compares strongly surviving NZ98/254 (serotype B:4:P1.7-2,4) and poorly surviving H34 (serotype W:nt:P1.18-3).
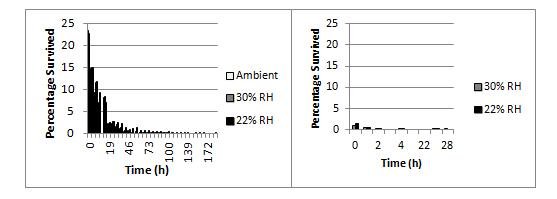
Figure 1. Effects of humidity on survival of N. meningitidis strains at 30 oC NZ98/254 (left panel) had >2% survival after 24 h when dried onto glass cover slips but H34 (right panel) survived for less than 4 h.
A quantitative proteomics workflow using iTRAQ labelling of proteins from N. meningitidis strains followed by protein identification using liquid chromatography tandem mass spectrometry was used to compare the protein profiles of strains under different environmental conditions (varying humidity). Figure 2 shows a COG (Clusters of Orthologous Groups) functional classification of proteins detected in the mass spectrometry analysis, including ribosomal subunits and elongation factors; membrane porins porA, porB, and rmpM; cell division proteins minD and ftsZ; glycolysis proteins gap, eno, ppsA, and fba; energy production proteins ackA1 (acetate kinase 1), pdhA (pyruvate dehydrogenase), gltA (citrate synthase), sucA and sucB (2-oxoglutarate dehydrogenase components), sucC and sucD (succinyl co-A ligase subunits), atpA and atpD (atp synthase subunits), lpdA1 (dihydrolipoyl dehydrogenase), icd (isocitrate dehydrogenase), and acnB (aconitate hydratase 2); pilin proteins pilQ and pilE; and protein-folding chaperones fkpA, dnaK, tig, slyD, groS, and groL.
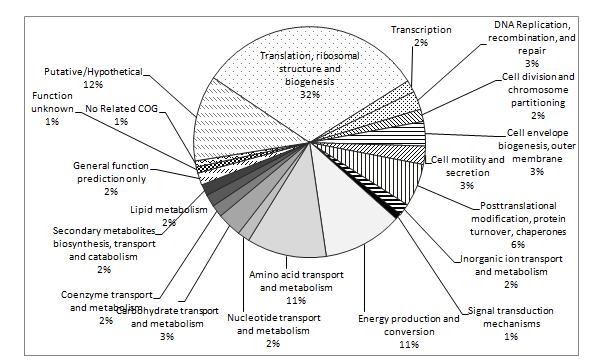
Figure 2. Functional classification of NZ98/254 proteins identified by LC-MALDI-MS/MS. The largest class identified were involved in translation and ribosomal structure, followed by energy production and amino acid metabolism. Twelve percent of the identified proteins were putative/hypothetical so this work was the first demonstration of their expression.
When grown at 30°C and 30% relative humidity, strain NZ98/254 produced significantly more porB (an anion-selective porin) and opa (an adhesion protein), than when grown under normal conditions (36°C, 5% CO2 and 95% RH). It also showed general increases in proteins associated with the membrane, amino acid biosynthesis, and some but not all stress-associated proteins. This same strain grown at 30°C and 22% relative humidity exhibited similar behaviour with some significant increases in amino acid biosynthesis (potD3), and membrane proteins (NMV_1429 and NMBNZ0533_1464, both of unknown function). This strain also showed a general increase in proteins involved in energy production and ribosomal proteins. In comparison, H34 showed no significant protein differences when grown at 30°C, 30% relative humidity than normal, but a general reduction in proteins involved in energy production, amino acid biosynthesis, and lipid metabolism was observed. These same general differences were observed at 22% relative humidity with a significant decrease in energy production proteins ppa and acnB, and despite an increase in amino acid biosynthesis associated potD3. The only conspicuous difference between the 30% relative humidity and 22% relative humidity behaviour was an increase in ribosomal protein expression at 22% relative humidity. The proteomic study is a first step towards identification of bacterial genes and proteins that are associated with environmental survival.



