Reports of research work funded by grants prior to 2016
Institute of Environmental Science & Research Ltd
Validation of White Blood Cell Lineage Specific DNA methylation markers
MC Benton, A Cameron, J Clapham, D Macartney-Coxson
Using publicly available data we identified a series of genes with cell specific DNA methylation that differentiates between white blood cell types. We hypothesised that cell specific DNA methylation influences the gene expression profiles of cell populations in different manners. Our project aimed to investigate epigenetic regulation of cellular lineage via the potential effects on cell specific mRNA expression profiles.
The original application which was funded by WMRF in 2015 had 4 main stages:
- a) sample (blood) collection and cell sorting (completed);
- b) pyrosequencing of identified DNA methylation markers (completed);
- c) QRTPCR to investigate mRNA expression of these genes (completed);
- d) Analysis and integration of the data (to be completed late 2016)
Our successful application was granted in May, starting mid-2015. At the time we proposed a 12 month period for completion. Due to several unforeseen delays we have not progressed as far as we would have hoped, however we believe we are now set up to complete this project before the end of 2016.
The first delay was due to the wait time on ethics approval. This was dependent on our application being heard and approved by the appropriate Health and Disability Ethics Committees, we could not collect samples before this time. Full ethics approval was obtained from the Health and Disability Ethics Committees on the 1 September 2015.
Patient recruitment, education and selection were performed during November 2015. Sample collection (blood) was performed at the Malaghan Institute of Medical Research by a trained medical professional during the month of December 2015. Twelve individuals fully consented and provided blood samples. Cell sorting is best performed on fresh samples, so individuals would provide samples in the morning and cell sorting of those samples was completed that same day. Cell sorting was performed at the Malaghan Institute of Medical Research by Alanna Cameron. We experienced some significant delays in progress due to the optimisation and implementation of the cell sorting phase. Upon completion of the cell sorting at the end of January 2016, all sample materials were transported to ESR Kenepuru Science Centre.
Pyrosequencing of bisulfite converted DNA was performed by EpigenDX, USA. Samples were shipped to EpigenDX late-March 2016. Assays were designed and optimised during April and data was fully received by mid-May 2016. Initial analysis of the methylation data looks extremely positive, with all cellular markers being positively identified (example Fig. 1).
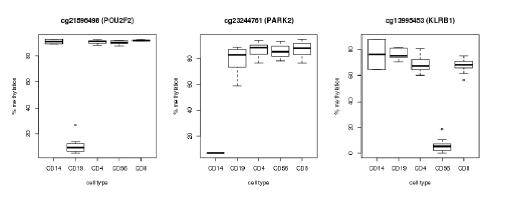 Figure 1 Preliminary analysis of the pyrosequencing data showing three methylation markers discriminating CD19, CD14 and CD56 cell types clearly from the other populations.
Figure 1 Preliminary analysis of the pyrosequencing data showing three methylation markers discriminating CD19, CD14 and CD56 cell types clearly from the other populations.
Real-time qtPCRs of selected genes was performed in house at ESR. This step of the project was completed at the end of May 2016.
With the recent completion of both data collection aspects we are now moving onto the analysis, integration and interpretation stage of this project. We anticipate that this should be a fairly straightforward task and hope to have all analysis work completed before the end of August 2016. Additionally, we aim to have a scientific manuscript submitted for peer-review by the end of the year (2016).



